Probabilistic Road Map based Protein Folding
CS365: Artifical Intelligence
Era Jain(Y9209)
Romil Gadia(Y9496)
Advisor: Prof. Amitabha Mukerjee
Abstract
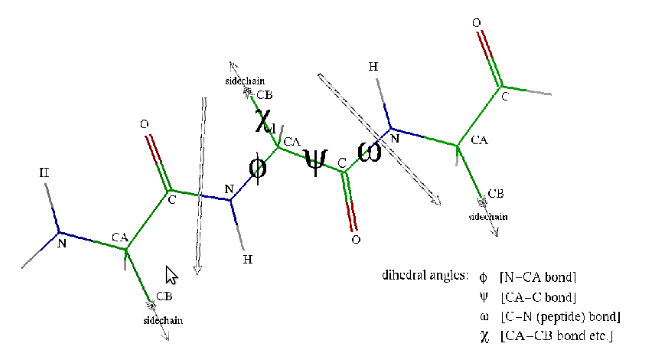
Protein Folding is a biochemical process but it is closely linked to Robotic Motion Planning. At first sight, robots and proteins seem to have little in common. But, they are very similar in their functionality which is based on their motions. Both, proteins and robots, can be modelled by their degrees of freedom(dof). This makes it possible to study motions in these two different domains by applying the same underlying algorithmic framework. Our method to study Protein Folding shall be derived from Probabilistic Road Maps(PRMs), originally developed for Robotic Motion Planning. The traditional methods of Motion Planning in protein are applied on the nodes in a Road Map, where the nodes represent the protein’s conformations. Our aim would be to create a smaller set of nodes which is approximate but representative of the protein’s energy landscape.
Links
Source Code
Project Report
Presentation
Project Proposal
Area
References:
[1]: A Motion Planning Approach to Studying Molecular Motions, Lydia Tapia, Shawna Thomas, Nancy M. Amato, Communications in Information and Systems, 10(1):53-68, 2010. Also, Technical Report, TR08-006, Parasol Laboratory, Department of Computer Science, Texas A&M University, Nov 2008.
[2]: Intelligent Motion Planning and Analysis with Probabilistic Roadmap Methods for the Study of Complex and High-Dimensional Motions, Lydia Tapia, Ph.D. Thesis, Parasol Laboratory, Department of Computer Science, Texas A&M University, College Station, Texas, Dec 2009.
[3]: Using Motion Planning to Study Protein Folding Pathways, Guang Song, Nancy M. Amato, Journal of Computational Biology, 9(2):149-168, Nov 2002. Also, In Proc. Int. Conf. Comput. Molecular Biology (RECOMB), pp. 287-296, Apr 2001. Also, Technical Report, TR00-026, Parasol Laboratory, Department of Computer Science, Texas A&M University, Oct 2000.
[4]: Image Source
[5]: Code Sources:
a. Python PDB Tools
b. Crankite program suite by Alexei Podtelezhnikov, Ph.D., Computational Biochemist, Biophysicist, and Bioinformatician
