Introduction:
Proteins are an essential part of life. They are important for the immune system because they are able to recognize foreign invaders and send appropriate signals to stop the spreading of infection or disease. An important aspect of proteins is their misfolding. Every protein needs to take a lowest energy conformation in order to react and function properly. The motion they perform largely affects their behaviour and functionality. If it can't fold properly, then it is called a "misfold" which can be devastating for an organism. Diseases like Alzheimer's and Mad Cow's disease are consequences of protein misfold. Origin of protein misfold is unclear, thus prediction of their 3-D conformations is an important problem. Knowledge of stability and kinetics may help provide insight into how proteins may fold.Problem:
Given the amino acid sequence (primary structure) of a protein, can we effectively predict its most stable 3-D conformation and the pathway it may follow to reach that configuration. It has been shown that the amino acid sequence of a protein has all the required information to determine its native (most stable) 3-D conformation. There's no perfect solution yet for this problem because a protein may assume enormous amount of possible 3-D conformations. Thus, a naive way of predicting the structure would be highly computationally intensive which is not feasible. This problem is shown to be NP-Hard. But, in nature we observe that a protein may shift across various conformations within millionth of a second to reach its native state. Thus, nature somehow has a polynomial time algorithm which we have been unable to find until now. Levinthal's ParadoxRelated Work:
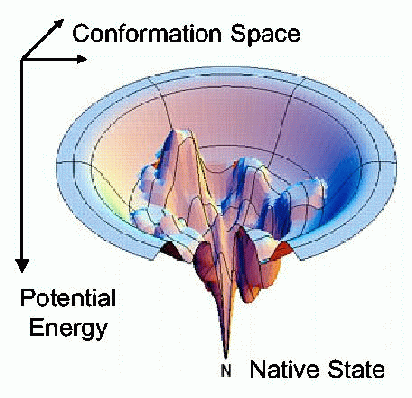
The protein folding phenomenon was largely an experimental problem until the formulation of the "Energy Landscape" theory of proteins by Joseph Bryngelson and Peter Wolynes in the late 1980s and early 1990s. An energy landscape is a mapping of all the conformations of a molecule to their respective Gibbs' free energy. This model (the energy landscape is a funnel with the native conformation at the bottom with the lowest energy) allows a protein to fold into its native state through a large number of pathways and intermediate states. Approaches like "Ab Initio", Genetic Algorithm and PRM (Probabilistic Road Map) have also been developed in the quest of an efficient algorithm.
Image Source: https://parasol.tamu.edu/foldingserver/FAQ_Technique.php
Our Ideas:
We plan to study two approaches viz., Probabilistic Road Map Method and the Genetic Algorithm approach. The former is inspired from the Motion Planning Approach used for robots. It uses the Funnel Energy Landscape Model. We aim to find some relationship among the two approaches, as if why do they converge to the same solution. We hope to find some pattern and constraints which both of them inadvertently satisfy in order to achieve the goal. The research papers we refer can be found in the References section below.Dataset:
https://parasol-http://www.cse.iitk.ac.in/users/cs365/2012/submissions/.cse.tamu.edu/groups/amatogroup/foldingserver/index.phpReferences:
1) Rufei Lu, Lauren Yarholar, Warren Yates, Dr. Miguel Bagajewicz, The University of Oklahoma [2008]: Protein Folding Prediction